Abstract
Background: Genetic studies have identified common variants within the HBS1L-MYB intergenic polymorphism (HMIP) region on chromosome 6q, BCL11A on chromosome 2p, and Xmn 1- HBG2 (rs748214) as associated with elevated fetal hemoglobin (HbF) levels and other clinically important human hematological traits. A recent study suggested that HbF is a predictor of outcome in myelodysplastic syndrome (MDS)/acute myeloid leukemia patients receiving decitabine. We set out to evaluate the genotypic significance of HbF and its effect on hematological traits in patients with myeloproliferative neoplasm (MPN), myeloma and MDS treated with HbF-inducing cytotoxic agents to determine potential of the HbF-associated variants for prediction of treatment response in these conditions.
Methods: 89 patients - 44 with MPN on Hydroxyurea (HU), 33 with myeloma on Lenalidomide, and 12 with MDS on Azacytidine were recruited from hematology clinics at Brighton & Sussex NHS Trust, UK. Blood samples were obtained from patients with written consent and analyzed for HbF/hematological parameters. 7 common HbF variants (rs6545816 and rs1427407 on BCL11A, rs9494142, rs6920211, rs9494145, and rs66650371 in HMIP, and rs748214) were genotyped on DNA extracted from peripheral blood using Taqman procedure. A subset of the patients (N = 16) (7 MPN, 6 myeloma and 3 MDS patients) had baseline values before starting therapy and had repeat values 3 months after cytotoxic agent initiation. The study was approved by the local and national UK NHS Research Ethics Committee (REC) (reference 14/WA/1078). Statistical analysis to test and graph genetic association of blood traits was performed with linear regression (IBM SPSS Statistics 24) with a genotypic genetic model adjusting for age and sex as covariates and analysis of variance determining genotype effect and mean HbF. Hardy-Weinberg test was performed on all genotype results for quality assurance. Hematological parameters which did not satisfy assumptions of normality were log-transformed.
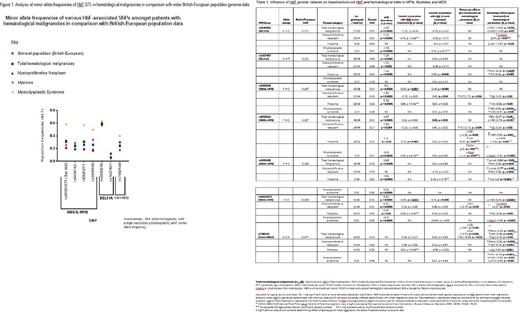
Results: HbF variants genotyped in our hematological malignancy study population had significantly discordant minor allele frequencies (MAF) differing in comparison to MAF from wider British/European population data derived from 1000 Genome project with MDS having greatest MAF discordance (see Fig. 1). Amongst the total hematological malignancy group, HMIP variants were statistically associated with higher baseline and induced HbF levels as well as lower WBC (see Table 1). Amongst MPN patients on HU, rs9494142 (HMIP) was observed as significantly associated with induced HbF levels (p = 0.04) in addition to increased baseline platelet count (p = 0.04) (see Table 1). These variants were seen to affect different hematological parameters at different stages of treatment as observed with HMIP variants rs9494142 and rs6920211 shifting from their regulation of baseline platelet levels to the regulation of induced hemoglobin levels (p = 0.02) 3 months post cytotoxic agent initiation. Amongst the Myeloma group, rs6920211( HMIP) was significantly associated with baseline HbF (p < 0.001). BCL11A variant rs1427407 was strongly associated with induced HbF levels with Lenalidomide (p = 0.002). rs1427407 was also significantly associated with increased platelet count (p = 0.04) negating the thrombocytopenic tendency of Lenalidomide (see Table 1). Xmn 1- HBG2 (rs748214) was not observed to have any statistically significant association with HbF but was associated with decreased hemoglobin (p = 0.03) in Myeloma and increased levels of hemolysis markers (reticulocytes (p = 0.003) and lactate dehydrogenase (p = 0.001)) in MPN patients on HU.
Conclusion: Although our sample size is small,the data suggests that the common HbF variants influence HbF and hematological traits in MDS, MPN, and myeloma and that the variants may potentially be involved in disease biology and treatment response in these conditions. The study also suggests HbF acts as a marker for the presence of the genetic variants that influences the hematological response in MDS/AML in the previous study. We hope that this study stimulates a larger prospective genotype-phenotype association studies of these disorders with the HbF variants.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal